I’ve been collaborating with Michael Simeone of I-CHASS on strategies for visualizing topic models. Michael is using d3.js to build interactive visualizations that are much nicer than what I show below, but since this problem is probably too big for one blog post I thought I might give a quick preview.
Basically the problem is this: How do you visualize a whole topic model? It’s easy to pull out a single topic and visualize it — as a word cloud, or as a frequency distribution over time. But it’s also risky to focus on a single topic, because in LDA, the boundaries between topics are ontologically sketchy.
After all, LDA will create as many topics as you ask it to. If you reduce that number, topics that were separate have to fuse; if you increase it, topics have to undergo fission. So it can be misleading to make a fuss about the fact that two discourses are or aren’t “included in the same topic.” (Ben Schmidt has blogged a nice example showing where this goes astray.) Instead we need to ask whether discourses are relatively near each other in the larger model.
But visualizing the larger model is tricky. The go-to strategy for something like this in digital humanities is usually a network graph. I have some questions about that strategy, but since examples are more fun than abstract skepticism, I should start by providing an illustration. The underlying topic model here was produced by LDA on the top 10k words in 872 volume-length documents. Then I produced a correlation matrix of topics against topics. Finally I created a network in Gephi by connecting topics that correlated strongly with each other (see the notes at the end for the exact algorithm). Topics were labeled with their single most salient word, except in three cases where I changed the label manually. The size of each node is roughly log-proportional to the number of tokens in the topic; nodes are colored to reflect the genre most prominent in each topic. (Since every genre is actually represented in every topic, this is only a rough and relative characterization.) Click through for a larger version.
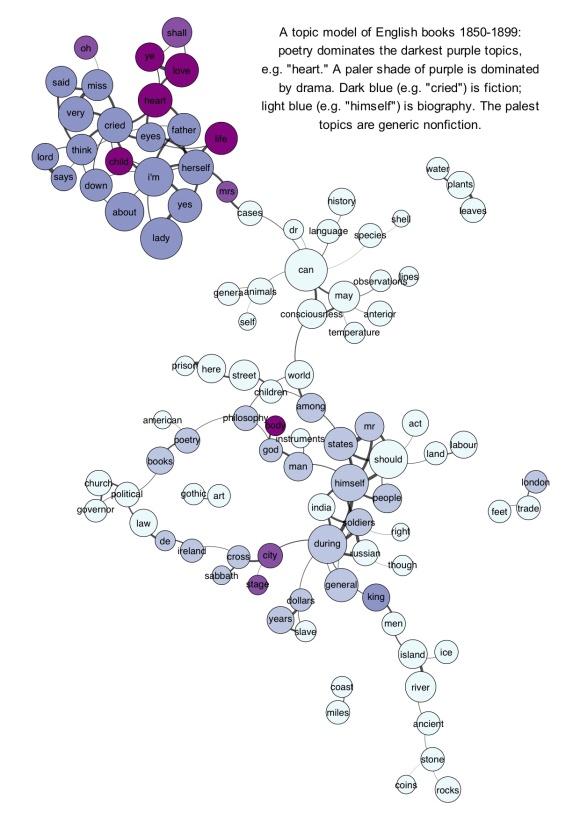
Since single-word labels are usually misleading, a graph like this would be more useful if you could mouseover a topic and get more information. E.g., the topic labeled “cases” (connecting the dark cluster at top to the rest of the graph) is actually “cases death dream case heard saw mother room time night impression.” (Added Nov 20: If you click through, I’ve now edited the underlying illustration as an image map so you get that information when you mouseover individual topics.)
A network graph does usefully dramatize several important things about the model. It reveals, for instance, that “literary” topics tend to be more strongly connected with each other than nonfiction topics (probably because topics dominated by nonfiction also tend to have a relatively specialized vocabulary).
On the other hand, I think a graph like this could easily be over-interpreted. Graphs are good models for structures that are really networks: i.e., structures with discrete nodes that may or may not be related to each other. But a topic model is not really a network. For one thing, as I was pointing out above, the boundaries between topics are at bottom arbitrary, so these nodes aren’t in reality very discrete. Also, in reality every topic is connected to every other. But as Scott Weingart has been pointing out, you usually have to cut edges to produce a network, and this means that you’re always losing some of the data. Every correlation below some threshold of significance will be lost.
That’s a nontrivial loss, because it’s not safe to assume that negative correlations between topics don’t matter. If two topics absolutely never occur together, that’s a meaningful relation! For instance, if language about the slave trade absolutely never occurred in books of poetry, that would tell us something about both discourses.
So I think we’ll also want to consider visualizing topic models through a strategy like PCA (Principal Component Analysis). Instead of simplifying the model by cutting selected edges, PCA basically “compresses” the whole model into two dimensions. That way you can include all of the data (even the evidence provided by negative correlations). When I perform PCA on the same 1850-99 model, I get this illustration. I’m afraid it’s difficult to read unless you click through and click again to magnify:
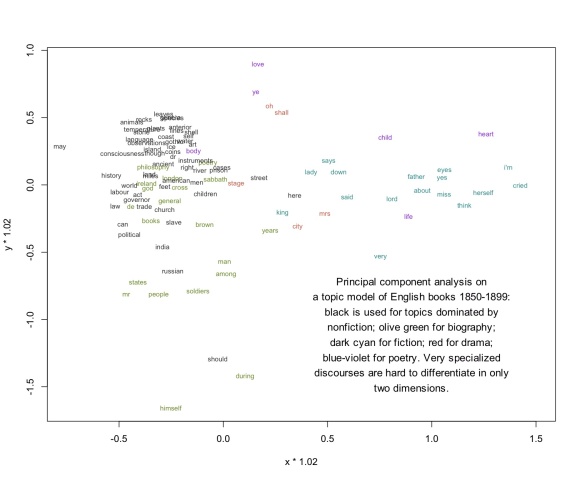
I think that’s a more accurate visualization of the relationship between topics, both because it rests on a sounder basis mathematically, and because I observe that in practice it does a good job of discriminating genres. But it’s not as fun as a network visually. Also, since specialized discourses are hard to differentiate in only two dimensions, specialized scientific topics (“temperature,” “anterior”) tend to clump in an unreadable electron cloud. But I’m hoping that Michael and I can find some technical fixes for that problem.
Technical notes: To turn a topic model into a correlation matrix, I simply use Pearson correlation to compare topic distributions over documents. I’ve tried other strategies: comparing distributions over the lexicon, for instance, or using cosine similarity instead of correlation.
The network illustration above was produced with Gephi. I selected edges with an ad-hoc algorithm: 1) take the strongest correlation for each topic 2) if the second-strongest correlation is stronger than .2, include that one too. 3) include additional edges if the correlation is stronger than .38. This algorithm is mathematically indefensible, but it produces pretty topic maps.
I find that it works best to perform PCA on the correlation matrix rather than the underlying word counts. Maybe in the future I’ll be able to explain why, but for now I’ll simply commend these lines of R code to readers who want to try it at home:
pca <- princomp(correlationmatrix)
x <- predict(pca)[,1]
y <- predict(pca)[,2]

30 replies on “Visualizing topic models.”
On word counts, I find usually find PCA discriminates better using log of counts as the input; might that also work with topic distributions?
If “closeness in space” is the important measure, hierarchical clustering of topics in text-space (or log topics) might be another good way to go: the first two principal components probably aren’t capturing all that much more information than nos. 3 and 4, and the two steps of correlating and then reducing dimensionality are probably doing a lot of work to ensure no especially surprising results come in.
Actually, that suggests an idea. The interesting thing about correlation space is that two topics that never appear in the same book could be right next to each other because they are used the same context. What would happen if you fitted some log-linear model to every pairwise distance in correlation space and in straight document space? Those might be very interesting relationships. My suspicion is that in those cases, the topic model is doing something we might call ‘failing,’ which would be good to now about.
Hierarchical clustering is a great idea! Acting on your suggestion, I just tried it in R (treating the topic x topic correlation matrix as a “distance” matrix), and the results make beautiful sense. I think that may actually be the best approach. It’s visually less pretty than PCA or network modeling, but I suspect it’s making better use of the data. The only problem is that it’s a bit hard to get 100-200 leaves to fit well on a single dendrogram. But maybe a visualization wizard can resolve that.
I had actually already tried PCA on log(wordcount + 1), because you’re right — the problem I ran into with raw wordcounts looked just like a logarithmic scaling problem. But for some reason, I can’t solve it by taking the log either before or after PCA. I may just be tripping on my shoelaces — not sure.
Well, there’s no strong reason to prefer the log raw counts to the correlation matrix in any case; it just might be interesting to see the differences. I’ll mark it down for later. Maybe try it on my ships.
It does seem like dendrograms have fallen behind other visualization techniques in the last few years; it’s too bad. If you’re in D3, it can do nice collapsible trees that could work pretty well if you’re willing to lose a bit of the distance information. I suppose hierarchical clustering is in theory another sort of network diagram (where you add nodes at the juncture points), so you could use almost any network visualization on it, too. Labelling the juncture-nodes would be tricky, though.
Oh, those collapsible trees are a very nice solution, because they could potentially be extended to include the top n words in each topic. That’s potentially the way to go — although, as you say, it gets tricky to label the juncture-nodes.
I also think hierarchical clustering is great for topic browsing. Especially compared to a flat list of the topics — you have to choose *some* ordering, so you might as well choose one where similar topics are next to each other, and might as well use a little bit of indentation while you’re at it. Here’s a really quick and dirty one I made a while ago, for a corpus of New York Times articles about crime.
Click to access crime.k=100.pdf
Top 8 or so words per topic. (The /N’s are because I preprocessed the corpus only to nouns; long story. And obviously it would be nice to not be backwards!)
It looks like I used Jensen-Shannon divergence between the topic-word probability vectors as the similarity metric, though I remember other things like cosine similarity may have been fine. (So this is a bit different than what you’re doing with doc-topic correlation matrix, though I think there’s some sort of perhaps-closed-form systematic mathematical relationship between these things.) I used R’s hclust() function… whatever the default is worked best if I remember right; I guess it’s complete-link clustering.
(Edit: looking at it more, obviously I did *not* filter to nouns; I guess all words are just annotated with the part-of-speech. Whatever, that’s irrelevant for this purpose)
Thanks! That looks good. I’ve used hclust() for some other things in the past, and it worked for this as well.But in the end I’m finding the dendrogram layout frustrating because it only really gives you one dimension. It works well if you’re interested in forming clusters, but I’m less interested in clustering here than in browsing the whole model. I do agree with you, though, that a hierarchically clustered listing would be better than a flat list in arbitrary order.
I’ve been doing lots of work clustering documents, as opposed to topics, but I’ve also found hierarchical clustering works well. It’s the basis of the document mining software I’m working on, Overview, which uses a collapsable tree-based visualization of the structure of the document vector space.
I would also recommend trying multi-dimensional scaling on the correlation matrix. mdscale() in R. This produces a low-dimensional projection like PCA but it’s non-linear and can capture cluster structure well. My experience is that it works reasonably well to give an idea of the high-dimensional structure of some set if neither the number of objects nor the sort of intrinsic dimension is too high. Otherwise you get featureless blobs (this paper for why.)
Nice. I just tried cmdscale() in R. It actually produces a layout that’s very, very close to the PCA layout. (Upside down, but otherwise almost the same.)
The “topic tree” function in Overview looks very nice, by the way!
Belated PS: I’ve got to admit, for the record, that the more I look at this problem, the more I’m drawn to the network solution. Even though the ostensible point of this post was to question it!
I still think the logical objections I raised in the post are cogent, and I think they should make us wary about applying the quantitative tools of network analysis to a topic “network.” But if you’re just looking for a casual “map” (and that’s really all I am looking for), the network viz. seems to have visual advantages that may outweigh its logical flaws.
A dendrogram, after all, only gives you one dimension. Even PCA only gives you two dimensions. But a network does a pretty good job of conveying the complexity of high-dimensional space (the way X and Y can be close to each other in one sense, although Y is close to Z in a completely different sense unrelated to X). Anyway, that’s what I’m thinking at the moment.
Be careful. You know that network graphs give you that extra dimensionality, but most who look at them will interpret Euclidean distance as topical distance.
I may not completely understand how you went from the initial text to PCA, but here are my questions:
– people usually try to interpret PCA dimensions – i.e., what is the meaning of X axis, and what is the meaning of Y axis, in terms of the data being used? Have you tried to do this?
– also, people often use PCA as a way to cluster data with similar properties together – in other words, typically in PCA the data points which are close to each other have something in common. However, in your application, you use already existing topic labels (shown via colors). In principle, a group such as the three words in the bottom of your PCA graph should have something in common (if I understand things correctly).
I have been using PCA extensively for couple of years, as I was also educating myself about details and proper use and interpetation – so its quite likely that my points are not correct – but may be worth thinking about.
Here is the best decription of PCA application (using R) I found:
http://factominer.free.fr/classical-methods/principal-components-analysis.html
Yes, you’re right that it’s worth interpreting dimensions. Broadly it looks to me like the first (x) component is something like (nonfiction vs. literature) and the (y) component is something like (science and nature at top / society and politics at bottom). Could confirm by checking the PCA components, but really, coloring topics by dominant genre makes the sorting pretty transparent.
Hello, everyone! Someone pointed to this thread of messages. What an interesting conversation! If you don’t mind me chiming in from the west coast…
The focus of analysis here seems to be about latent topics, where each topic is represented as a probability distribution over terms. Assuming the goal is to show the “whole model,” wouldn’t the most relevant component of an LDA model be the topic-term probability? If so, how about showing the probability distribution itself?
For example, here is an LDA model about concepts in information visualization.

The areas of the circles in the matrix are proportional to the probability of a term belonging to a topic. On the left is a list of the most salient terms (i.e., frequent and distinctive). On the top are the 50 latent topics. The bottom of the matrix is cut off from the screenshot, but one could scroll down to display more terms in the actual visualization.
The effectiveness of a matrix view depends heavily on the ordering of its rows and columns. Here, the words are re-ordered to cluster related terms and improve the readability of multi-word phrases. If you scan the word list starting from the top, you should be able to read off “online communities”, “social network(s)”, “node-link diagrams”, etc.
No attempt was made to re-order the topics. This view was originally generated to help evaluate the quality of individual latent topics. However, one could conceivably re-order the topics to reveal topical groupings.
Even with this screenshot, one could see that the orange topic (#25) overlaps with the purple topic (#41). Both share common vocabulary such as “layout algorithms” and “maintaining stability.” Purple is more about “force-directed graph layout” while orange deals more with “ordered treemap”, “favorable aspect ratio”, etc.
Hi Jason, I love that visualization (I had actually seen it or another like it somewhere), and it would work perfectly for the problem of characterizing relationships between a small number of closely-related topics. I can envision using it, once I had chopped off a subset of topics and a subset of vocabulary relevant to them. Then I might want to understand exactly how they interlock.
But I have a hard time imagining how it would work on a whole model of the size I want to generate. The model above is 100 topics and 10,000 words, and that’s actually sort of a “trial” or “sample” size version. Really what I want is more like 250 topics and 50,000 words. I don’t think it will be possible to differentiate a model of that scale on a grid like this. But it could be used for portions of the problem.
For instance, all the topics probably share certain common words, even though they don’t pop up as “salient” using Blei’s formula (that saliency formula, incidentally, seems to do a whole lot of the work of LDA though it rarely gets discussed!) We could use a grid visualization to see how the topics relate to each other in a limited subset of common words. Might be interesting.
As I watch this thread accumulate more and more responses, I keep thinking about it. So:
I think you’re selling short hclust on this: “But a network does a pretty good job of conveying the complexity of high-dimensional space (the way X and Y can be close to each other in one sense, although Y is close to Z in a completely different sense unrelated to X).” Although PCA is locked into certain dimensions.
This is right about PCA, but I think not about hierarchical clustering. You can force-direct a dendrogram, and then it’s just a particular sort of network graph in two dimensions that captures the same sort of twisted manifold of multi-dimensional space you’d want. And if you use ‘friend of friends’ clustering (‘single’ vs ‘complete’ in R’s hclust), X and Z don’t need to be close to each other. And it captures something about the layout that an adhoc strategy doesn’t: what the really weird outliers in the set are.
To my mind, the ideal network visualization would be to have edges weighted by some standard measure of distance between topics, and just let the whole field sort itself out. That seems in theory like the best way of getting as much higher-dimensional data as possible. But I bet it doesn’t work, because a) there aren’t that many great network placement algorithms for weighted edges that don’t produce a hairball. Using arbitrary cutoffs or clustering measures seem like similar tradeoffs.
Another algorithm that may be better than PCA for this sort of task is T-SNE, which (I think) can keep in the variable higher-dimensional relations better than PCA at the cost of absolute distances being meaningful. It seems best at computer vision problems and I can’t find any citations to network uses, but it seems like it might have some promise as a network-layout algorithm. Maybe nobody knows about it yet.
Thanks, Ben, I appreciate these additional suggestions. In fact I just went and tried hclust with the “single” method. (I was using “ward.”) On the dataset I’m working with now, “single” produced a tree with a strange structure — one very big cluster without a lot of internal structure and a couple of very small ones. But I’ll keep fiddling.
My default assumption now is that it’s good to have a lot of arrows in your quiver. I think PCA tells you something, and so does hclust. Probably good to use several methods, if you really want to understand the model.
I think you’re probably right that we need better force-directed algorithms. For instance, in this particular problem, there are negative correlations between topics. So it might be useful to have “negative” edges — connections between two nodes that actually operate as repulsions rather than attractions. That might do something to loosen up the “hairball” effect you otherwise get if you include all edges.
Right now I’m experimenting with a network graph that is also an image map — so by clicking on a node you go to a page with a lot of information about that node that’s unrepresentable in the overall graph. This is the solution I’m happiest with so far for browsing. But I suspect there’s not going to be any one solution, because browsing is one goal, clustering is another, understanding PCA space is another …
Huh, now that I think about it, that makes sense for single: I suppose it’s basically reproducing the single cluster you get with the network analysis, except without the internal ordering (because doesn’t tell you which point in the cluster it was closer to). So that’s pretty much useless, except as a commentary that in one important sense there aren’t distinct groups in the set. Which isn’t much help when the whole point is to look at whatever distinct groups there might be in the set.
What placement algorithm are you using, by the way? One of the weird things about network science is that while you have to justify PCA, Kamada-Kawai or whatever is somehow more naturalized.
When Facebook was very young, one of the basic features was a social-network graph of your friends. The FAQ answered the question “What placement algorithm are you using for the social graph?” with something along the lines of “shut up, nerd.” Because the graphs just direct themselves, you know.
The “single” hclust did separate out a few topics that are actually in German or French. (Was topic modeling a journal where some articles are not in English.) So in one sense it was capturing the most important high-level structure. But …
I’m building networks in Gephi, and using whatever algorithm they call “Force Atlas 2.” Plus the rule “prevent overlapping nodes.” And I agree with you about the spurious transparency of network-building. It involves so many judgment calls. In Gephi you can sort of drag nodes around while the placement algorithm is working, to encourage a more legible structure, which is great fun, very useful, and completely unjustifiable.
E.g. in the network above, you’ll notice that “water-plants-leaves” are up by the other science topics. Why? No edges connect them. Two separate networks. I just dragged ’em up there because they seemed sciencey. 🙂
[…] Underwood’s most recent suggested solution seems to be folding in Principal Component Analysis, and that’s where I’m headed next. […]
[…] DADA manifesto of 1921). What digital analysts are doing with text using tools (see Ted Underwood, “Visualizing Topic Models”), the dadaists were doing by hand using […]
I have just started learning about topic modelling and found that Mallet tool works good for my purpose. But I was wondering about how to visualize it in gephi. In this blog post you do explain how you created the gephi file. But I am not very familiar with implementing algorithms. So could you please share the code/script to convert topic modelling result -> correlation matrix -> gephi node/edge table? It would be of great help. Thanks!
I’m new to topic modeling and text mining, but with some background in econometrics and basic stats. In getting started on text mining, I’ve been surprised by the lack of basic correlation matrices, or corrgrams (http://www.datavis.ca/papers/corrgram.pdf), or heatmaps showing the relationships between the underlying word frequencies. Instead, most papers jump quickly to clusters or PCA. I wonder if this doesn’t make dimension reduction more mysterious than it needs to be, especially to humanists without a strong math background.
[…] made out of the DADA manifesto of 1921). What digital analysts are doing with text using tools (see Ted Underwood, “Visualizing Topic Models”), the dadaists were doing by hand using […]
I wonder how your results would compare to those of a correlated topic model, which is designed specifically to accommodate how topics are related to one another: http://www.cs.princeton.edu/~blei/papers/BleiLafferty2007.pdf
‘topicmodels’ has a CTM function, and integrates well with other text mining / natural language processing packages to clean up the data prior to analysis. however, it apparently lags behind the ‘mallet’ package in terms of extracting data for later use in network analysis.
[…] “Visualizing Topic Models” by Ted Underwood https://tedunderwood.com/2012/11/11/visualizing-topic-models […]
Construction loans work as interest only mortgages for the
duration of the construction phase. This offers the lender the ability to better decide whether it is a right conclusion to lend to that individual
or not. If you are reading this and no one has
passed on, the best way to prepare for a funeral for
a loved one is to either take out a guaranteed life insurance policy, enough to cover funeral expenses, or start a savings plan to cover funeral expenses.
[…] Ted Underwood’s: Visualizing topic modells […]
[…] deal with this problem, Ted Underwood came up with a really clever link-cutting heuristic that produces much cleaner network diagrams. However, it’s a little […]
1
1
11